Practical application of cell counting method performance evaluation and comparison derived from the ISO Cell Counting Standards Part 1 and 2
Cell & Gene Therapy Insights 2021; 7(9), 937–960
10.18609/cgti.2021.126
The increased utilization of cells in biomanufacturing and as therapeutic products over the last decade has prompted the development and publication of two ISO Cell Counting Standards, ISO 20391 – 1:2018 and ISO 20391 – 2:2019 to provide guidance on general principles relating to cell counting and to establish an approach to evaluate the quality of cell counting methods. In this work, we demonstrate the practical implementation of the experimental protocol outlined in ISO Cell Counting Standard Part 2 and a Bland-Altman comparative analysis to evaluate performance and comparison of cell counting methods. We compare two cell types, two image cytometry instruments, and two fluorescent stains, calculating the precision, coefficient of determination (R2), and a proportionality index (PI) parameter to evaluate cell counting method performance. In addition, the cell counting results are directly compared to evaluate bias between two cell counting methods. The protocol is suitable for evaluating and comparing the performance of multiple cell counting methods to select for downstream assays.
Introduction
In the recent decade, cell and gene therapies have drastically improved their efficacy and have become essential players in cancer treatment [1]Boyiadzis MM, Dhodapkar MV, Brentjens RJ et al. Chimeric antigen receptor (CAR) T therapies for the treatment of hematologic malignancies: clinical perspective and significance. J. ImmunoTher. Cancer 2018; 6(137): 1–12., [2]Gonçalves GAR, Paiva RdMA. Gene therapy: advances, challenges and perspectives. Einstein 2017; 15(3): 369–75.. With the approval of two chimeric antigen receptor (CAR) T cell therapies by the U.S. Food and Drug Administration (FDA) in 2017, the numbers of clinical studies and tests on new and novel cell therapy products have also surged [3]Huang R, Li X, He Y et al. Recent advances in CAR-T cell engineering. J. Hematol. Oncol. 2020; 13(86): 1–19., [4]Li Y, Huo Y, Yu L, Wang J. Quality Control and Nonclinical Research on CAR-T Cell Products: General Principles and Key Issues. Engineering 2019; 5: 122–31., [5]Seimetz D, Heller K, Richter J. Approval of First CAR-Ts: Have we Solved all Hurdles for ATMPs? Cell Med. 2019; 11: 1–16.. Typically, cellular therapies require genetic modification of the immune cells (i.e. T cells, NK cells) collected from patients, culture expansion, and re-introduction of the final products back into the patients. Therefore, it is critical to provide accurate cell counting for the administration of proper dosages, which may otherwise lead to inefficacy or induce unwanted autoimmune responses in patients undergoing therapeutic treatments [6]Nagata S, Hanayama R, Kawane K. Autoimmunity and the Clearance of Dead Cells. Cell 2010; 140(5): 619–30., [7]Rock KL, Kono H. The inflammatory response to cell death. Ann. Rev. Pathol. 2008; 3: 99–126., [8]Zhang Y, Chen X, Gueydan C, Han J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018; 28: 9–21..
In the 21st Century Cures Act, the United States Congress has also recognized the importance of standardization for streamlining development, quality assurance, and facilitating regulatory approval of cell and gene therapy products [9]. In the “Synergizing Efforts in Standards Development for Cellular Therapies and Regenerative Medicine Products” workshop held by the FDA on March 31st, 2014, cell counting and viability measurement assurance were identified as opportunities for standards development [10]Arcidiacono JA, Bauer SR, Kaplan DS, Allocca CM, Sarkar S, Lin-Gibson S. FDA and NIST collaboration on standards development activities supporting innovation and translation of regenerative medicine products. Cytotherapy 2018; 20: 779–84., [11]Lin-Gibson S, Sarkar S, Elliott JT. Summary of the National Institute of Standards and Technology and US Food And Drug Administration cell counting workshop: Sharing practices in cell counting measurements. Cytotherapy 2018; 20(6): 785–95.. ISO has since published two cell counting standards, “ISO 20391-1:2018 Biotechnology – Cell Counting – Part 1: General Guidance on Cell Counting Methods” and “ISO 20391-2:2019 Biotechnology – Cell Counting – Part 2: Experimental Design and Statistical Analysis to Quantify Counting Method Performance”, which can serve as guidance for researchers working in the field of immunotherapy and adoptive cell therapy, where both require high quality and robust cell counting measurements for biologics and cell products [12]Biotechnology – Cell counting – Part 1: General guidance on cell counting methods. In: Standardization IOf, editor; 2018., [13]Biotechnology – Cell counting – Part 2: Experimental design and statistical analysis to quantify counting method performance. In: Standardization IOf, editor; 2019..
Derived from general concepts described in ISO Cell Counting Standard Part 1, we propose 6 key factors that can provide guidance on the selection of cell counting methods and improve the quality of the cell counting measurements:
- Determine the intended use of the cell counting result (e.g. cell count for normalization of bioassays, cell therapy dosing, post-tumor digestion for single cell-based transcriptome analysis, mouse tissue processing for cytotoxicity assays, or isolation of human PBMCs for immunophenotyping analysis, etc.)
- Investigate to understand cell sample composition (e.g. various cell types, particle debris, chemical impurities, and suspension medium), as well as the morphological appearances of the cells under microscopy
- Understand the assay principles and select the appropriate cell counting assay, such as total, live and dead cell count, viability, or cell population analysis
- Investigate the capabilities and select the appropriate cell counting systems, where the system consists of reagents, consumables, instrument, and software algorithms, as well as assay performance criteria (i.e. precision, range, linearity, etc.)
- Treat each cell counting method as a whole process, including sampling, diluting, and staining, which are critical for proper sample preparation
- Provide continuous operator training, in order to ensure consistent cell counting results
It is also realized that the cell counting needs for cell and gene therapies are broad due to a wide range of biological sample types with various formulations and bioprocessing steps, which are complex, dynamic, and heterogeneous. Because there are currently no reference materials for live mammalian cells that are certified for cell concentration, the accuracy parameter outlined in the ICH Harmonised Tripartite Guideline – Validation of Analytical Procedures: Text and Methodology Q2 (R1) cannot be readily applied, thus increasing the challenge and difficulty of validating the accuracy of cell counting [14]Validation of Analytical Procedures: Text and Methodology Q2(R1). In: Guideline IHT, editor; 2005., [15]Guidance for Industry: Process Validation: General Principles and Practices. In: Services USDoHaH, Administration FaD, (CDER) CfDEaR, (CBER) CfBEaR, (CVM) CfVM, editors; 2011., [16]Guidance for Industry: Analytical Procedures and Methods Validation for Drugs and Biologics. In: Services USDoHaH, Administration FaD, (CDER) CfDEaR, (CBER) CfBEaR, editors; 2015.. Therefore, the ISO cell counting standards can serve as a valuable tool to evaluate and select cell counting methods that are fit-for-purpose, in order to increase confidence in the cell counting results.
Development of the protocol
We have employed the guidance from ISO 20391 – 2:2019 Biotechnology – Cell Counting – Part 2 and utilized information from the ICH Q2 (R1) to develop an appropriate protocol to evaluate the performance of selected cell counting methods. The ICH Q2 (R1) guidance document presents multiple parameters for the validation of analytical methods, such as robustness, linearity, detection range, limits of detection (LOD), limits of quantitation (LOQ), precision (repeatability, intermediate precision, reproducibility), and accuracy. It is important to note that since there is no reference material to provide a reference value for cell concentration, the evaluation of the accuracy parameter needs to be indirectly assessed by orthogonal comparative methods. The ISO Cell Counting Standard Part 2 describes a detailed protocol to simultaneously evaluate precision (repeatability), coefficient of determination (R2), and proportionality.
Utilizing the ISO Cell Counting Standard Part 2 document, we have identified several key parameters that can quickly and sufficiently assess the performance of cell counting methods [17]Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21., [18]Sarkar S, Pierce L, Lin-Gibson S, Lund SP. Standards Landscape in Cell Counting: Implications for Cell & Gene Therapy. Cell Gene Ther. Ins. 2019; 5(1): 117–31.Sarkar S, Pierce L, Lin-Gibson S, Lund SP. Standards Landscape in Cell Counting: Implications for Cell & Gene Therapy. Cell Gene Ther. Ins. 2019; 5(1): 117–31.. In this work, we will focus on an experimental protocol derived from the ISO Cell Counting Standard Part 2, which evaluates the coefficient of determination (R2[CV)], [and proportionality index (]PI) of a cell counting method. The proportionality index is a metric introduced in the ISO Cell Counting Standard Part 2 that quantifies the degree to which a cell counting method conforms to the principle of proportionality, where it is expected that cell counts will scale proportionally with dilution. The principle of proportionality is a fundamental property of any cell counting method, and any deviation from proportionality would indicate a systematic or non-systematic error resulting in a loss of measurement accuracy. To more directly evaluate systematic deviation from proportionality, which is an indicator of loss of accuracy, the PI is calculated by fitting a proportional model to the dilution series data, then summarizing residuals based on smoothed data, thus reducing the influence of random variation on the evaluation of proportionality [17]Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.. There are several approaches to calculate PI, where some PI metrics may be more relevant based on the fit-for-purpose need of the cell counting method. Some metrics penalize more for outliers, while others weigh errors evenly across the dilutions or allow more contribution by higher cell concentrations. In this work, we utilized the PI model published previously from the National Institute of Standards and Technology (NIST) [17]Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.. It should be noted that sources of systematic error which are proportional to sample dilution will not be detected with this approach. For example, if debris are mixed with the cell suspension and falsely identified as cells, concentration of both cells and debris would be proportionally reduced with dilution, and the false counts would not affect the proportionality. In order to demonstrate the appropriate usage of the proposed experiments, these protocols were tested using various image cytometry systems from Nexcelom Bioscience LLC. (Lawrence, MA).
Bland-Altman comparative analysis method
Comparative analysis methods can be employed to compare the performance of different cell counting methods. While the lack of reference material precludes the direct measurement of cell counting accuracy, comparison of orthogonal methods may serve as a viable alternative. It is also often desirable to determine how closely the results of one method will agree with another, such as when an instrument is upgraded after many years in the lab. One useful method is the construction of a Tukey mean-difference plot, also known as a Bland-Altman plot [20]Altman DG, Bland JM. Measurement in Medicine: The Analysis of Method Comparison Studies. J. Royal Stat. Soc. Series D (The Statistician) 1983; 32(3): 307–17., [21]Bland JM, Altman DG. Statistical Methods For Assessing Agreement Between Two Methods of Clinical Measurement. Lancet 1986; 327(8476): 307–10., [22]Tholudur A, Giron L, Alam K et al. Comparing Automated and Manual Cell Counts for Cell Culture Applications. BioProcess Int. 2006.. The Bland-Altman analysis results in the calculation of a bias (with corresponding confidence interval) between two methods, indicating which method counts higher or lower on average and by how much. The analysis also provides an estimate of how well the two methods are expected to agree for a single sample. Here we modify the dilution series experimental design described in ISO Cell Counting Standard Part 2 document to collect data appropriate for a Bland-Altman analysis while also meeting the standards requirements for calculating CV, R2, and PI.
Usually, Bland-Altman plots consist of absolute differences between two measurements plotted against their mean. In the case of cell counting, variance is not constant for different concentrations, but is generally proportional to the number of cells counted [17]Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.. For Bland-Altman analysis to be useful in such an application, the data can be transformed to achieve roughly constant variance across a range of concentrations. In this protocol, we use percent differences rather than absolute differences to achieve more uniform variance.
The Bland-Altman analysis method produces three metrics of comparison:
- The bias between two methods, which is the mean of the differences.
- The limits of agreement (LoA), which are a multiple of the standard deviation of the differences.
- The confidence interval (CI) of the bias, which is a multiple of the standard error of the mean of the differences.
The bias describes the average difference between measurement results obtained via the two methods. Due to biological variation in the samples and variability in the measurement process for both methods, it is impossible to predict exactly how much the measurement of any single sample will differ between the two methods. However, when measurements of many samples are averaged, a bias – even a slight one – may become clear. The bias may be interpreted as one method measuring higher or lower than another on average, though the difference between methods when measuring a single sample may vary widely.
The limits of agreement describe how widely these differences may vary. When added to and subtracted from the bias, the LoA define a range within which the difference between the measurements from two methods of a single sample is expected to be found. In this protocol, we use the limits of agreement that approximate a 95% confidence interval (1.96 × standard deviations) for a normal distribution. This is a sufficient approximation for our purposes and 72-measurement sample size. If fewer measurements are acquired, confidence intervals calculated from the appropriate t distribution (rather than the Normal distribution) are advised. If the percent differences between the results from the two methods follow a normal distribution, we can expect that 95% of the differences will fall within one LoA from the value of the bias. In reality, the values will not be strictly normal, but the approximation is useful for evaluating subsequent measurements [23]Carkeet A. A Review of the Use of Confidence Intervals for Bland-Altman Limits of Agreement in Optometry and Vision Science. Optometry Vision Sci. 2020; 97(1): 3–8.. If more statistical rigor is required, tests for normality can be applied, and the confidence intervals can be more exactly calculated [24]Carkeet A. Exact Parametric Confidence Intervals for Bland-Altman Limits of Agreement. Optometry Vision Sci. 2015; 92(3): e71–e80.. Depending on the variation observed between samples relative to the variation between replicate measurements from each sample, it may be helpful to include random effects terms typically included in analysis of hierarchical experiments. In this work, we were not concerned with the sample-to-sample variation in the proposed experiments.
The confidence interval of the bias provides the approximate uncertainty for the calculated bias value and suggests a range within which the true value of the bias between the two cell counting methods is likely to be found. Unlike the LoA, this confidence interval narrows with an increased number of samples measured. If the 95% confidence interval is larger than the absolute value of the bias (i.e. the CI brackets the value 0), the method comparison has not demonstrated a statistically significant bias between the two methods (at α = 0.05 significance level). With enough samples, even a very slight bias may be confidently measured. A slight bias is often negligible compared to sample variation. Researchers should consider how a cell count is being used in order to determine acceptable levels of bias in their case.
Before proceeding with Bland-Altman comparative analysis for cell counting, researchers should:
- Determine the range of cell concentration values for which comparison between the two methods is desired;
- Determine what values of the bias and LoA are acceptable for their application,
- Select cell samples that are representative of the population for which the comparison is desired and the range determined in step 1; and
- Measure each sample using the different cell counting methods, taking care that the sample does not change between measurements (minimal delay between measurements, proper mixing, etc.) [25]Abu-Arafeh A, Jordan H, Drummond G. Reporting of method comparison studies: a review of advice, an assessment of current practice, and specific suggestions for future reports. Br. J. Anaesthesia 2016; 117(5): 569–75..
A higher number of paired measurements can reduce the uncertainties of the bias and LoA, e.g. the confidence interval of the bias narrows with more measurements. Researchers should determine the precision they require, and increase the number of paired measurements accordingly – we suggest a minimum of 20 paired measurements be used as a starting point. Finally, it is possible that either the bias or the variation will vary with cell concentration. In such a case, the bias and LoA obtained for the entire group of data may not be representative of how the two methods compare over a narrower range of concentrations. It may be useful to perform Bland-Altman analysis on smaller subsets of data.
Applications of the method
The cell counting method performance evaluation and comparison protocols can be applied to research, analytical method development, process development, and preclinical or clinical trials. In addition, the method can be applied to a plethora of research fields requiring the usage of cells such as cellular and gene therapy, immuno-oncology and immunotherapy, cell line development and biologics production, virology and infectious disease, regenerative medicine, toxicology, food science, and even renewable energy. The quality of cell counting results is critical for a wide range of cell types used in the research fields mentioned above. These cell types can include primary cells such as human or mouse whole blood, cord blood, bone marrow aspirate, adipose tissue, hepatocytes, PBMCs, leukapheresis sample, platelets, tumor or tissue digests are typically used. In addition, bacteria and yeast cells are often used to generate biologics or used for beverage production.
Experimental design
The cell counting method performance evaluation proposed here consists of a dilution series experiment and comparative analysis for multiple methods. The experimental design is demonstrated using CHO-S and Jurkat cell lines fluorescently stained with acridine orange and a green nuclear dye. Two cell counting systems are compared: the Cellaca MX High-Throughput Cell Counter (Cellaca MX) and the Celigo Image Cytometer (Celigo). It is important to note that ISO Cell Counting Part 2 requires users to assess pipetting error contributions to dilution integrity to establish confidence in dilution and sampling. Here, we conducted a pre-evaluation of pipetting error, which will not be described in this protocol. It is also important to investigate the stability of the target cell sample prior to conducting the experiment in order to avoid drift in concentration and viability during the assay time frame. The stability of the Jurkat and CHO cells used in this work have been previously tested and showed no noticeable trends (Supplementary Figure 1).
The dilution experiment consists of a 6-point concentration series of the target cell types, where each concentration is independently produced from the original stock (rather than the other dilutions) to reduce a propagation of dilution error that can affect proportionality. The dilution series should span the typical concentration range of the target cell samples in order to evaluate the performance of the cell counting method in the specified range.
Three replicate samples are generated per concentration, and each replicate sample is measured 4 times per cell counting method so that each method provides a total of 12 measurements per concentration and a total of 72 measurements in a 6-point concentration series. The measurements are used to calculate the coefficient of determination (R2), precision (repeatability – Coefficient of Variation, CV), and proportionality index (PI) parameters for each cell counting method. It is important to note that the tested Jurkat and CHO cells were stained with acridine orange and Nuclear Green dye to measure only the total cell concentration in this work.
For performance comparison between two cell counting methods, the Bland-Altman method is applied. Like the proportionality measurement, the comparison results are valid only for the intended use of the specific methods (cell type, assay type, exact instruments, etc.), and only for the range of cell concentrations included in the test. Therefore, it is vital to first define the exact methods, test conditions, and range of cell concentrations over which comparison is desired. For most accurate results, the Bland-Altman analysis should include as many measurements as possible, encompassing the sources of variation that are expected for the normal operation of the cell counting method, such as multiple operators, reagent lots, and cell culture flasks. Each point on the Bland-Altman plot is obtained by using both cell counting methods to measure a single sample. The sample should be carefully mixed to ensure homogeneity before portions are taken for measurement with each cell counting method. Measurements should be made with minimal lag time between them, simultaneously if possible. If the experiment described above is performed with the same tubes of cells using both methods at the same time, Bland-Altman analysis may be performed with the resulting data. If desired, tighter confidence intervals on the calculated bias or less uncertainty on the Limits of Agreement can be obtained by supplementing the data with more samples. Concentrations spanning the selected concentration range should be represented roughly equally in the samples used.
Expertise needed to implement the protocol
In general, the expertise required to implement the cell counting method performance evaluation is proper training by an expert user in the operation of the cell counting systems. In addition, the users should be trained on sample preparation to ensure consistent performance of the dilution, sampling, and staining steps of the cell counting process.
Limitations
Accuracy is one of the most critical parameters for the validation of an analytical method, however, it cannot be directly applied to most cell counting methods. Since there are limited live cell reference standards, it is challenging to assess the accuracy of a cell counting method. Therefore, proportionality is an alternative parameter to assess accuracy relative to dilution fraction, which serves as the internal control, as well as utilizing orthogonal methods for comparison.
It should be recognized that R2 values calculated over a range of concentrations are strongly dependent on the range chosen. A larger range of linear data results in an R2 value closer to 1. If comparison between R2 values is to be attempted, it is important that the range for the two calculations be the same. In addition, it should be noted that the proportionality index as defined here is not normalized to the number of dilution fractions and the number of biological replicates per dilution fraction. It is required that the same experimental design be used if PI is to be meaningfully compared between methods.
Materials
Documentation materials
- ISO 20391-1:2018 Biotechnology – Cell Counting – Part 1: General Guidance on Cell Counting Methods
- ISO 20391-2:2019 Biotechnology – Cell Counting – Part 2: Experimental Design and Statistical Analysis to Quantify Counting Method Performance
Biological materials
- Chinese Hamster Ovary (CHO-S) cell line (Gibco, #11619012)
- Jurkat, Clone E6-1 cell line (ATCC, TIB-152™)
Growth medium & supplements
- CD CHO Medium (1X) (Gibco, #10743011)
- GlutaMAX-1 (100X) (Gibco, #35050061)
- HT Supplement (100X) (Gibco, #11067030)
- RPMI Medium 1640 (1X) (Gibco, #11875093)
- Fetal Bovine Serum (FBS) (Access, #A19023)
- Antibiotic Antimycotic Solution (100X) (Sigma-Aldrich, #A5955-100ML)
Fluorescent staining reagents
- ViaStain™ AOPI Staining Solution (AOPI, Nexcelom Bioscience, CS2-0106-5mL)
- ViaStain™ AO Staining Solution (AO, Nexcelom Bioscience, CS2-0108-5mL)
- ViaStain™ Total Cell Nuclear Green (Nuclear Green, Nexcelom Bioscience, CS1-V0008-1)
Other reagents & chemicals
- Phosphate Buffered Saline (PBS) powder (Sigma-Aldrich, #P38135)
- HyClone™ Water, Cell Culture Grade (Endotoxin-Free) (GE Health, #SH3052903)
Equipment
- Tissue culture hood (Forma Scientific, ClassII A/B3 BSC)
- Cell culture incubator (Thermo, Forma 370)
- Plate rocker (Boekel, RockerII 260350)
- Automatic pipettor (Fisherbrand™ Pipet Controller, #FB14955202)
- Manual pipettors (P10, P100, P1000) (VWR, 1-10UL, 10-100UL, 100-1000UL)
- Centrifuge (Eppendorf, 2702)
- Cellometer Spectrum and operating laptop computer (Spectrum, Nexcelom Bioscience)
- Cellaca MX High-Throughput Automated Cell Counter and operating laptop computer, concentration range of 1 x 105 – 1 x 107 cells/mL (Nexcelom Bioscience)
- Celigo Image Cytometer and operating desktop computer (Nexcelom Bioscience)
Disposable instruments
- T-75 cm2 flask (USA Scientific, CC7682-48)
- 15-mL centrifuge tube (Greiner Bio, 188271)
- Serological Pipets 5 mL, 10 mL, 25 mL (USA Scientific, #1075-0110, #1071-0810, #1072-5410)
- Pipette tips (P10 and P1000) (VWR, 7320561, 83007-380)
- Pipette tips (P200) (USA Scientific, 11111210)
- Microtubes 1.5 mL (VWR, 89000028)
- Microtubes 0.5 mL (CellTreat, 229440)
- Cell counting slides (Nexcelom Bioscience, CHT4-SD100-002)
- Cellaca MX High-throughput Automated Cell Counter Plates (Cellaca MX plates, Nexcelom Bioscience, CHM24-A100-001)
Reagent setup
Phosphate-Buffered Saline (PBS) solution
Prepare the PBS solution by mixing 5 L of H2O with 1 packet of PBS powder to generate a solution of 0.01M PBS at pH 7.4 with NaCl at 0.138 M and KCl at 0.0027 M.
CHO-S cell culture medium
Prepare CHO-S medium (500 mL) with the CD CHO Medium (1X) and supplement with 5 mL of the GlutaMAX-1 (100X) and 5 mL of the HT Supplement (100X).
Jurkat cell culture medium
Prepare Jurkat medium (500 mL) with the RPMI Medium 1640 (1X) and supplement with 10% FBS (50 mL) and 5 mL of the Antibiotic Antimycotic Solution (100X).
ViaStain™ AOPI Staining Solution
The acridine orange (AO) and propidium iodide (PI) staining solution is already prepared to the correct concentration before staining at 1:1 with the cells.
ViaStain™ AO Staining Solution
The acridine orange (AO) staining solution is already prepared to the correct concentration before staining 1:1 with the cells.
ViaStain™ Total Cell Nuclear Green staining solution
Prepare a 2X staining solution (10 µM) by mixing PBS and the Nuclear Green stock solution at 5 mM. Pipette 10 mL of PBS into a 15-mL centrifuge tube and add 20 µL of the Total Cell Nuclear Green stock solution. Close the 15-mL centrifuge tube and invert 10X to mix the staining solution before use.
Equipment setup
Cellometer Spectrum
Connect the Cellometer Spectrum to the operating laptop computer via the USB cable and plug in the power cord. Turn the instrument power on from the back side and then open the Cellometer Spectrum analysis software. In the Cellometer Spectrum software, select the “AOPI Viability Assay_S5” default assay type for cell counting.
Cellaca™ MX High-Throughput Automated Cell Counter
Connect the Cellaca MX to the operating laptop computer via the USB cable and plug in the power cord. Turn the instrument power on from the back side and then open the Cellaca MX analysis software. In the Cellaca MX software (v1.2), select the “MX04.0_AOPI_LiveDead” default assay type for cell counting.
Celigo® Image Cytometer
Turn on the Celigo power on the front and open the Celigo analysis software. Navigate to the top right and click on ‘Administration’ and then select ‘Manage Plate Profiles’. After the “Plate Profile Management” window opens, click on the ‘Import’ button and select the plate profile for Cellaca 12 × 2 plate. Return to the home screen for image acquisition and analysis.
Procedure
Maintenance of CHO-S cells
Timing: 20 – 30 min for passaging the cells and measuring their concentration and viability.
- Passage the CHO-S cells when they are between 2 to 4 × 106 cells/mL. Allowing the cells to grow above that concentration may decrease cell division as well as decrease viability due to insufficient nutrients in the media.
- Warm the CHO-S cell culture medium at 37°C for 15 min in the incubator or in a water bath at 37°C for 5 min before passaging.
- Under the biosafety cabinet, use a 10 mL pipette, pipette up and down at least 10 times to break up the cell clumps, and create a homogenous cell suspension in the T-75 flask.
- Remove 200 µL of cells from the T-75 flask and transfer into a 1.5 mL microtube before the cells have had a chance to settle.
- Obtain a CHT4-SD100 cell counting slide and peel off the protective plastic film on the top and bottom, and place the slide on a Kim-Wipe.
- Mix 20 µL of CHO-S cell sample and 20 µL of AOPI within a 0.5 mL microtube.
- Pipette 20 µL of stained cell sample into one chamber on the cell counting slide.
- Insert the cell counting slide into the Spectrum and select the “AOPI Viability Assay_S5”.
- Measure the cell concentration and viability.
- Based on the measured concentration, calculate the ratio of cells to new media that is needed in order to achieve a concentration of 2 × 105 cells/mL.
- Remove the calculated cell volume from the flask and replace with an appropriate amount of warmed CHO-S cell culture medium.
- Place the passaged flask back onto the plate rocker inside the 8% CO2 incubator at 37°C.
- Monitor the growth of cells daily, and continue to passage as needed (usually 3 times a week).
Maintenance of Jurkat cells
Timing: 20–30 min for passaging the cells and measuring their concentration and viability.
- Passage the Jurkat cells when they are between 1 to 2 × 106 cells/mL. Allowing the cells to grow above that concentration may decrease cell division as well as decrease viability due to insufficient nutrients in the media.
- Warm the Jurkat cell culture medium at 37°C for 15 min in the incubator or in a water bath at 37°C for 5 min before passaging.
- Under the biosafety cabinet, use a 10 mL pipette, pipette up and down at least 10 times to break up the cell clumps, and create a homogenous cell suspension in the T-75 flask.
- Remove 200 µL of cells from the T-75 flask and transfer into a 1.5 mL microtube before the cells have had a chance to settle.
- Obtain a CHT4-SD100 cell counting slide, peel off the protective plastic film on the top and bottom, and place the slide on a Kim-Wipe.
- Mix 20 µL of Jurkat cell sample and 20 µL of AOPI within a 0.5 mL microtube.
- Pipette 20 µL of stained cell sample into one chamber on the cell counting slide.
- Insert the cell counting slide into the Spectrum and select the “AOPI Viability Assay_S5”.
- Measure the cell concentration and viability.
- Based on the measured concentration, calculate the ratio of cells to new media that is needed in order to achieve a concentration of 2 × 105 cells/mL.
- Remove the calculated cell volume from the flask and replace with an appropriate amount of warmed Jurkat cell culture medium.
- Place the passaged flask back inside the 5% CO2 incubator at 37°C.
- Monitor the growth of cells daily, and continue to passage as needed (usually 3 times a week).
Stock cell sample preparation from cell culture
Timing: 15 min for collecting the cells from cell culture flasks, 5 min for cell counting and viability analysis, and 10 min for adjusting cell sample concentration if necessary.
- Collect a stock of CHO-S and Jurkat cell sample separately into a 15-mL tube from cell culture following aseptic techniques.
- Obtain a CHT4-SD100 cell counting slide, peel off the protective plastic film on the top and bottom, and place the slide on a Kim-Wipe.
- Pipette 20 µL of the cell sample using a P100 pipettor into a 0.5 mL microtube.
- Pipette 20 µL of the AOPI and add to the 0.5 mL microtube.
- Aspirate the mixture of cells and AOPI up and down at least 5 times.
- Pipette 20 µL of the stained cells into one chamber on the cell counting slide.
- Insert the cell counting slide into the Spectrum and count the stained cells to generate cell count and viability.
- Adjust the stock cell sample concentration to ~5 × 106cells/mL for both CHO-S and Jurkat cells.
- Decrease the concentration by dilution in cell media.
- Increase the concentration by centrifugation and resuspend in cell media.
- Repeat steps 28–33 to ensure the concentration is adjusted to ~5 × 106 cells/mL.
Sample preparation & cell counting preparation for cell counting methods performance evaluation & comparison
Timing: 15–30 min with a single, manual pipette for sample preparation. 15–20 min for incubation of cell samples mixed with Nuclear Green (Figure 1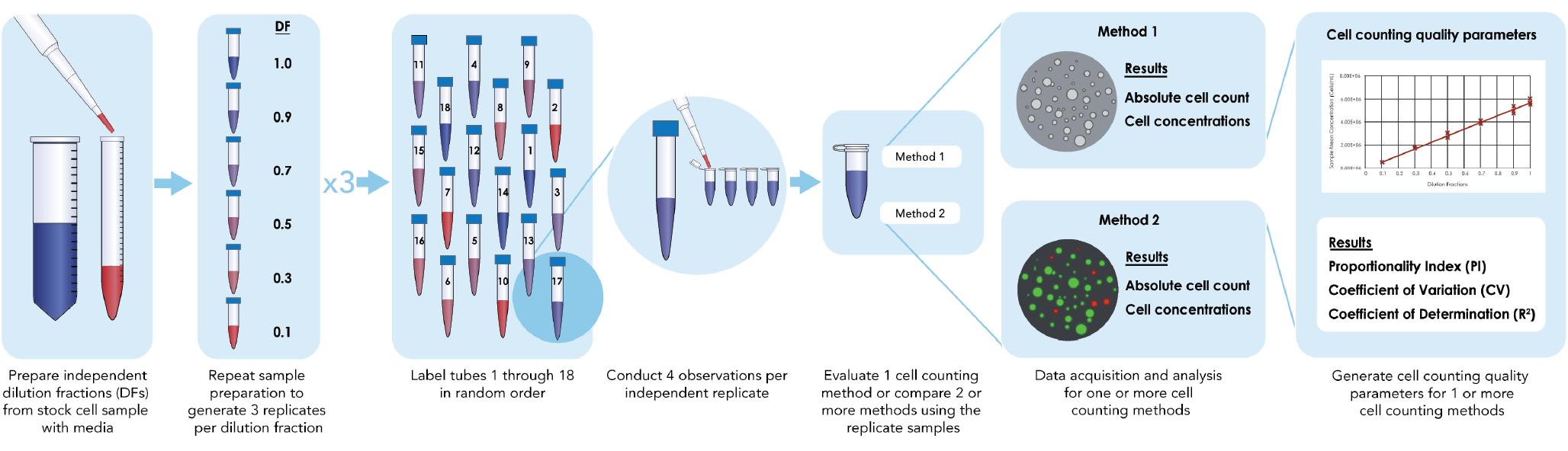
CRITICAL: Under the guidance of ISO Cell Counting Part 2, an initial accuracy validation experiment of pipetting volume using the experimental pipettors is necessary to increase sampling confidence. Such validation can be performed with a sensitive and well-calibrated laboratory balance and a fluid of known density, but the procedure will not be described in this protocol. Directly dilute the cells to generate independent dilution samples instead of serial dilution to reduce the propagation of pipetting error that can affect proportionality.
- Obtain the prepared stock CHO-S cell and Jurkat cell samples at the highest concentration for the intended use and range (~5 × 106 cells/mL).
- Prepare other samples from the stock of CHO-S and Jurkat cell samples at 0.1, 0.3, 0.5, 0.7, 0.9 and 1.0 dilution fractions (DFs) independently (Table 1).
- Prepare replicate samples with PBS or cell culture media.
| Table 1 Dilution fractions and the corresponding volumes preparation for cell sample and PBS. | ||
| DF | Cell volume (µL) | PBS volume (µL) |
| 1.0 | 120 | 0 |
| 0.9 | 108 | 12 |
| 0.7 | 84 | 36 |
| 0.5 | 60 | 60 |
| 0.3 | 36 | 84 |
| 0.1 | 12 | 108 |
- Pipette 120 µL of CHO-S or Jurkat stock cell sample into the 1st microtube for the 1.0 DF sample.
- Pipette 108 µL of CHO-S or Jurkat stock cell sample into the 2nd microtube and add 12 µL of PBS for the 0.9 DF sample.
- Pipette 84 µL of CHO-S or Jurkat stock cell sample into the 3rd microtube and add 36 µL of PBS for the 0.7 DF sample.
- Pipette 60 µL of CHO-S or Jurkat stock cell sample into the 4th microtube and add 60 µL of PBS for the 0.5 DF sample.
- Pipette 36 µL of CHO-S or Jurkat stock cell sample into the 5th microtube and add 84 µL of PBS for the 0.3 DF sample.
- Pipette 12 µL of CHO-S or Jurkat stock cell sample into the 6th microtube and add 108 µL of PBS for the 0.1 DF sample.
- Repeat Steps 38–43 two more times to generate a total of 3 replicate samples at each DF, where a total of 18 tubes of cell samples are generated.
- Pipette 120 µL of AO staining solution into the 1st microtube of each DF sample to make a 1:1 mixed sample. After this step, a total of 240 µL cell sample is prepared in the 1st microtube at each DF.
- Invert the 0.1 DF microtube 10 times to ensure uniform mixture.
- Transfer 50 µL from the mixed 0.1 DF microtube into the A1 loading well on the 1st Cellaca MX plate. Repeat the transfer 3 more times into the A2 – A4 loading wells of the 1st Cellaca MX plate.
- Repeat Step 46–47 for the 1st microtubes of the remaining DFs (0.3, 0.5, 0.7, 0.9, and 1.0) samples into the remaining loading wells on the 1st Cellaca MX plate, following the plate map shown below. After this step, the 1st Cellaca MX plate is prepared (Table 2).
- Randomize Step 46–48 if applicable. This is suggested by ISO Cell Counting Standard Part 2 in order to minimize the systematic time-dependence effects on the proportionality index and other metrics of the cell counting measurement process quality.
| Table 2 Cellaca plate map for cell samples at different DFs. | ||||||||||||
| Plate 1 | ||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | 0.3 | 0.3 | 0.3 | 0.5 | 0.5 | 0.5 | 0.5 |
| B | 0.7 | 0.7 | 0.7 | 0.7 | 0.9 | 0.9 | 0.9 | 0.9 | 1.0 | 1.0 | 1.0 | 1.0 |
| Plate 2 | ||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | 0.3 | 0.3 | 0.3 | 0.5 | 0.5 | 0.5 | 0.5 |
| B | 0.7 | 0.7 | 0.7 | 0.7 | 0.9 | 0.9 | 0.9 | 0.9 | 1.0 | 1.0 | 1.0 | 1.0 |
| Plate 3 | ||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 | 0.3 | 0.3 | 0.3 | 0.5 | 0.5 | 0.5 | 0.5 |
| B | 0.7 | 0.7 | 0.7 | 0.7 | 0.9 | 0.9 | 0.9 | 0.9 | 1.0 | 1.0 | 1.0 | 1.0 |
- Repeat Step 45–48 for the 2nd and 3rd replicate samples at different DFs to prepare the 2nd and 3rd Cellaca MX plates.
- Prepare each Cellaca MX plate right before the image acquisition, instead of preparing all Cellaca MX plates at the beginning, to minimize the time gap between sample preparation and image acquisition.
- Repeat Steps 36–49 and stain with 120 µL of Nuclear Green.
- Incubate the Nuclear Green-stained cell samples for 15–20 min at room temperature. Incubation time can be reduced at 37 °C.
Image acquisition & analysis for each cell counting method
Timing: Scanning and analysis are 6 min per plate for the Cellaca MX and 5–10 min per plate for the Celigo.
- Load the 1st Cellaca MX plate into the Cellaca MX after preparation.
- Select the “MX04.0_AOPI_LiveDead” default assay type for cell counting in the Cellaca MX software for cell samples stained with AO staining solution. For cell samples stained with Nuclear Green, increase the FL1 exposure time by 50–100%. Check the fluorescent intensity of the nuclear green stained cells in the preview images before image acquisition.
- Use the default analysis parameters for counting cells in the captured Cellaca MX bright field and FL1 fluorescent images. Export the concentration data.
- Transfer the 1st Cellaca MX plate to the Celigo. Select the plate profile for Cellaca MX plates. Use the default experiment setting for image acquisition and analysis. Export total cell counts from the captured fluorescence images.
- Repeat 51–54 for the 2nd and 3rd Cellaca MX plates.
Cell counting method performance evaluation
Timing: ∼30 min to calculate and analyze the parameters for performance evaluation for one cell line and one stain.
- Calculate the cell concentration using the total cell counts from the Celigo exported data and multiply by a factor of 1383.979, which is the conversion ratio based on the counted volume and dilution factor from staining with AO and Nuclear Green.
- Calculate the mean concentration MAi acquired with method A (Cellaca MX) from a total number of nAi replicate measurements for sample i using Equation 1.
[1]
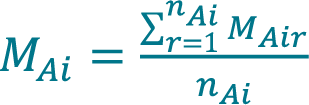 , where MAir is the concentration acquired with method A for sample i during replicate measurement r.
, where MAir is the concentration acquired with method A for sample i during replicate measurement r.
- Calculate the mean concentration MAk acquired with method A (Cellaca MX) for dilution fraction k (DFk) using Equation 2.
[2]
 Calculate the variance of concentration varAi acquired with method A (Cellaca MX) from a total number of nAi replicate measurements for sample i using Equation 3.
Calculate the variance of concentration varAi acquired with method A (Cellaca MX) from a total number of nAi replicate measurements for sample i using Equation 3.
[3]
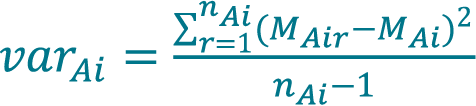 Calculate the pooled variance of concentration varAK acquired with method A (Cellaca MX) for DFk using Equation 4.
Calculate the pooled variance of concentration varAK acquired with method A (Cellaca MX) for DFk using Equation 4.
[4]
 Calculate the pooled standard deviation of concentration σAk acquired with method A (Cellaca MX) for DFk using Equation 5.
Calculate the pooled standard deviation of concentration σAk acquired with method A (Cellaca MX) for DFk using Equation 5.
[5]
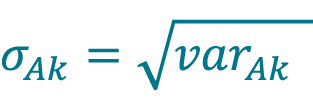 Calculate the pooled CV, CVAk acquired with method A (Cellaca MX) for DFk using equation Equation 6.
Calculate the pooled CV, CVAk acquired with method A (Cellaca MX) for DFk using equation Equation 6.
[6]
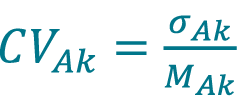 Repeat 57–62 to calculate the mean concentration MBk, the pooled standard deviation of concentration σBk and the pooled CV acquired with method B (Celigo) for DFk.
Repeat 57–62 to calculate the mean concentration MBk, the pooled standard deviation of concentration σBk and the pooled CV acquired with method B (Celigo) for DFk.
- Use mean concentrations (MAi, MBi) from all samples at 6 different DFs to generate a concentration series for both Cellaca MX and Celigo. Perform a proportional fit with the concentration series for each method using the iteratively reweighted least squares (IRLS) model. Set the weights of the least squares proportional to the reciprocal of the variances, which can be estimated by mean concentrations under the assumption of a quasi-Poisson distribution that the variances of cell concentrations are proportional to their respective mean concentrations (varAi = φMAi), where φ is a scalar estimated from the experimental data that cancels out when used in the weighting of every least squares term [17]Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.. Re-run the model fitting by updating weights using predicted values of the mean concentrations until the proportional fit is optimized. Generate a list of predicted values of mean concentrations (M̂Ai,ideal, M̂Bi,ideal) from the IRLS model.
- Determine the coefficient of determination (R2 value) from the IRLS model for method A (Cellaca MX) using Equation 7[26]Navidi W. Statistics for Engineers and Scientists. New York: McGraw-Hill; 2013., [27]Sokal RR, Rohlf FJ. Biometry. New York: W. H. Freeman and Company; 1969.. Use the same method to determine the R2 value for method B (Celigo).
[7]
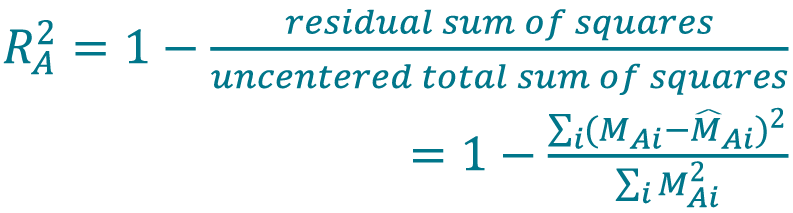 Perform a fit with the concentration series for each method (Cellaca MX, Celigo) using a higher-order polynomial model as a flexible model. Set the order of the polynomial to be the number of DFs minus 1. Generate a list of predicted values of mean concentrations (M̂Ai,flex, M̂Bi,flex) from the polynomial model.
Perform a fit with the concentration series for each method (Cellaca MX, Celigo) using a higher-order polynomial model as a flexible model. Set the order of the polynomial to be the number of DFs minus 1. Generate a list of predicted values of mean concentrations (M̂Ai,flex, M̂Bi,flex) from the polynomial model.- Determine the proportionality index (PI) based on the smoothed sum of absolute scaled residuals (PIASAbsSR, PIASAbsSR) for both Cellaca MX and Celigo using Equation 8 following previous publication [17]Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21.Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21., [18]Sarkar S, Pierce L, Lin-Gibson S, Lund SP. Standards Landscape in Cell Counting: Implications for Cell & Gene Therapy. Cell Gene Ther. Ins. 2019; 5(1): 117–31.Sarkar S, Pierce L, Lin-Gibson S, Lund SP. Standards Landscape in Cell Counting: Implications for Cell & Gene Therapy. Cell Gene Ther. Ins. 2019; 5(1): 117–31.,
[8]
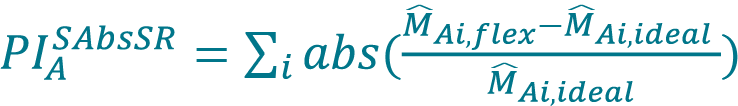
- Apply the Bland-Altman method to compare the performance between two cell counting methods.
- We utilized an internally developed software application derived from the ISO Cell Counting Standard Part 2 and Bland-Altman comparative method to automatically calculate the coefficient of determination, precision, proportionality index parameters, as well as the Bland-Altman analysis parameters (bias, LoA, the CI of the bias).
Bland-Altman comparative method: data calculation
Timing: ∼30 min to analyze and plot the Bland-Altman comparison data for one cell line and one stain.
- Calculate the percent difference Yi between the measurement MAi acquired with method A and the measurement MBi acquired with method B for each sample i using the Equation 9, only if the samples are paired between method A and B.
[9]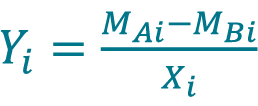 , where Xi is the sample mean given by
, where Xi is the sample mean given by 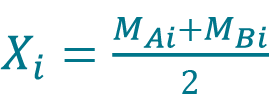
- If measurements MAir and MBir from replicate r are paired, calculate the percent difference Yir between the measurement MAir acquired with method A and the measurement MBir acquired with method B for each replicate r of sample i using the Equation 10.
[10]
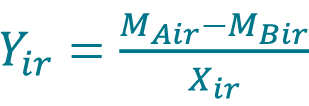 , where Xir is the sample mean given by
, where Xir is the sample mean given by 
Calculate the bias from method A to method B (BiasAB) by averaging the Yi values using Equation 11 or by averaging the Yir values using Equation 12
[11]
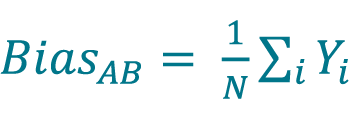 , where N is the number of samples (for paired samples, unpaired replicates, i.e. for each sample, different replicates are measured with each method).
, where N is the number of samples (for paired samples, unpaired replicates, i.e. for each sample, different replicates are measured with each method).
[12]
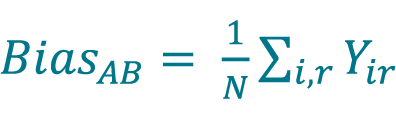 , where N is the total number of replicate measurements (for paired samples with paired replicates, i.e. for each sample, the same replicates are measured using both methods).
, where N is the total number of replicate measurements (for paired samples with paired replicates, i.e. for each sample, the same replicates are measured using both methods).
- Calculate the LoA by multiplying 1.96 to the mean for percent differences determined in step 70 using Equation 13 or 14. LoA are defined as the one-sided 95% confidence interval for a single sample.
[13]
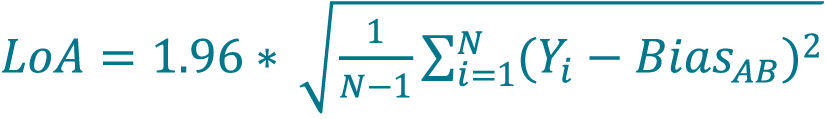
where N is the number of samples (paired samples, unpaired replicates).
[14]
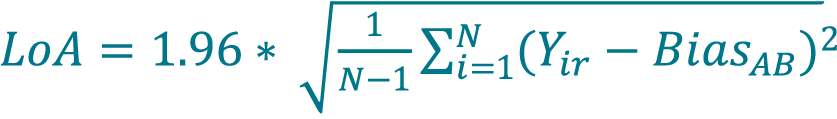
where N is the total number of replicate measurements (paired samples, unpaired replicates).
- Calculate the CI of the bias using Equation 15.
[15]

, where N is the number of samples (for paired samples without paired replicates) or the total number of replicate measurements (if both samples and replicates are paired).
Bland-Altman comparative method: graphical representation
- Plot a single point on the Bland-Altman diagram for each sample, with Xi (sample mean) on the horizontal axis and Yi (percent difference) on the vertical axis.
- Plot a horizontal line that crosses the vertical axis at the value of BiasAB calculated in step 71.
- Plot two additional horizontal lines that cross the vertical axis at the values of BiasAB + LoA and BiasAB – LoA, where the LoA is calculated as described in step 72. These lines define a range of values for the expected percent difference between the two methods for a single sample.
- Plot two additional horizontal lines at the values BiasAB + CIBias and BiasAB – CIBias. This range provides a sense of the uncertainty on the bias value itself.
- Examine the plot and note any concentration-dependence in either the bias or variation.
Troubleshooting
Follow the troubleshooting Table 3 to optimize the experiments and output.
| Table 3 Troubleshooting table. | |||
| Step | Problem | Possible reason | Solution |
| 62, 63 | CV is too large at one or a few DFs | Sampling or pipetting error |
|
| Counting errors due to clumps |
| ||
| 67 | Poor Proportionality | Propagation of pipetting error |
|
| 68, 70–78 | A large bias between two cell counting methods | Sample variation (i.e. different stocks of samples) |
|
| Sample condition change (e.g. photobleaching, sample dry-out) |
| ||
| Cell counting analysis variation (e.g. declumping) |
| ||
| Instrument comparison |
| ||
| Instrument calibration |
| ||
Timing
- Step 1–26, maintenance of CHO-S and Jurkat cells: 20–30 min for passaging the cells and measuring their concentration and viability per cell line.
- Steps 27–35, stock cell sample preparation from cell culture: 15 min for collecting the cells from cell culture flasks, 5 min for cell counting and viability analysis, and 10 min for adjusting cell sample concentration if necessary.
- Steps 36–50, sample preparation and cell counting preparation for cell counting methods performance evaluation and comparison: 15–30 min with a single, manual pipette for sample preparation, 15–20 min for incubation of cell samples mixed with Nuclear Green, and
- 0 min per Cellaca MX plate for cell counting preparation.
- Step 51–55, image acquisition and analysis: 6 min per plate for the Cellaca MX and 5–10 min per plate for the Celigo.
- Step 56–78, performance evaluation and Bland-Altman comparison analysis: 1 h per cell line per staining solution.
Anticipated results & discussion
Two cell lines (CHO-S, Jurkat), two dyes (AO, Nuclear Green), and 2 cell counting methods were evaluated to demonstrate the application of cell counting method performance evaluation and Bland-Altman comparative analysis. Figure 2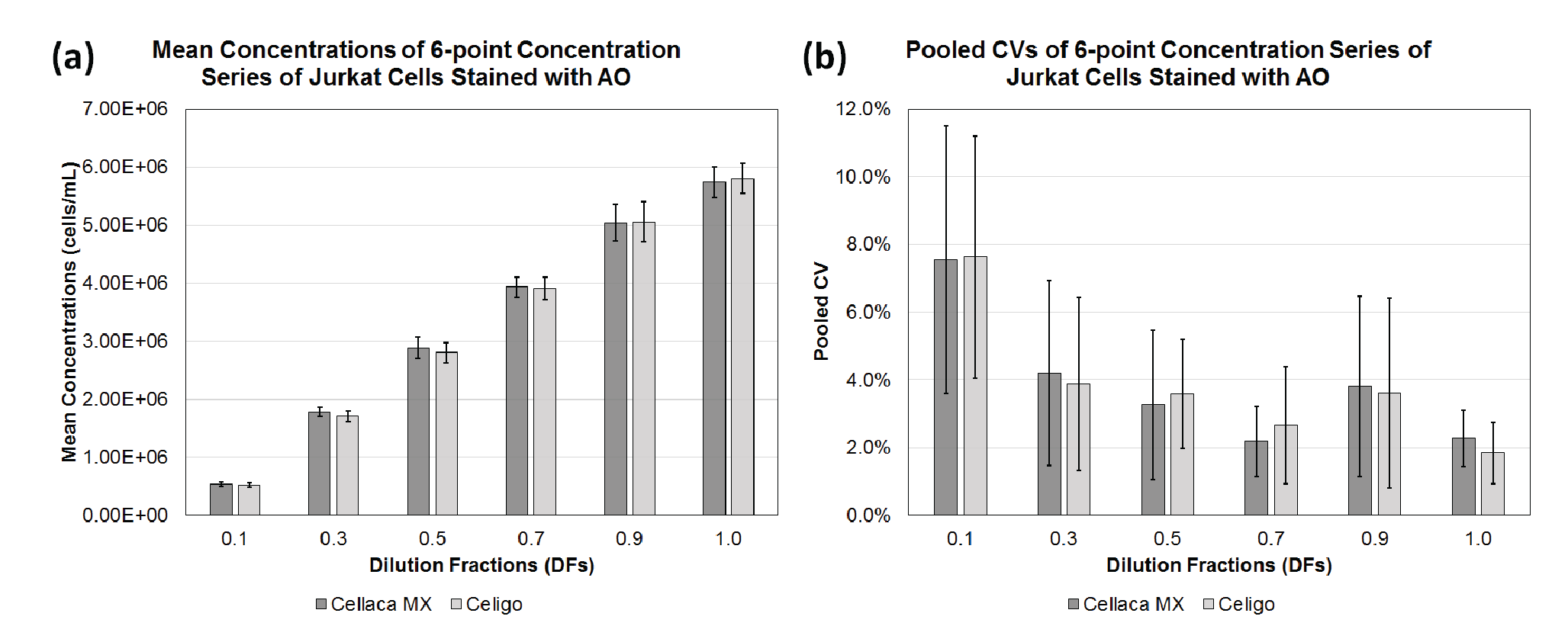
| Table 4 Calculated mean concentration and pooled CV for each dilution fraction using ISO Cell Counting Standard Part 2. | ||||
| Cell counting method | DF | n | Mean (cells/mL) | Pooled CV (%) |
| Cellaca MX | 0.1 | 12 | 5.37E+05 | 7.5 |
| 0.3 | 12 | 1.78E+06 | 4.2 | |
| 0.5 | 12 | 2.88E+06 | 3.3 | |
| 0.7 | 12 | 3.93E+06 | 2.2 | |
| 0.9 | 12 | 5.05E+06 | 3.8 | |
| 1.0 | 12 | 5.74E+06 | 2.3 | |
| Celigo | 0.1 | 12 | 5.21E+05 | 7.6 |
| 0.3 | 12 | 1.71E+06 | 3.9 | |
| 0.5 | 12 | 2.80E+06 | 3.6 | |
| 0.7 | 12 | 3.91E+06 | 2.7 | |
| 0.9 | 12 | 5.06E+06 | 3.6 | |
| 1.0 | 12 | 5.80E+06 | 1.8 | |
Based on the measurements in Table 4, the coefficient of determination and proportionality index can be calculated via regression analysis. Figure 3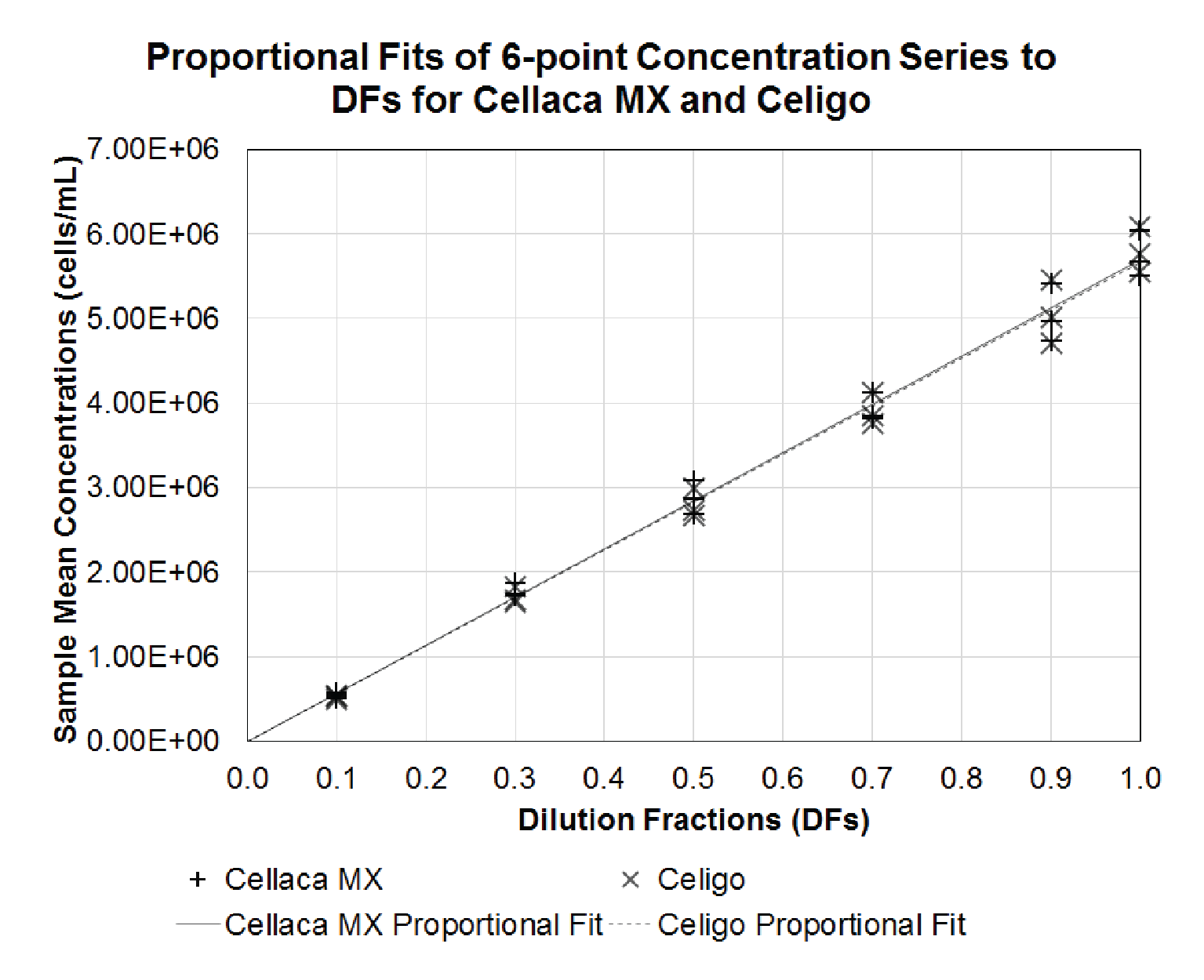
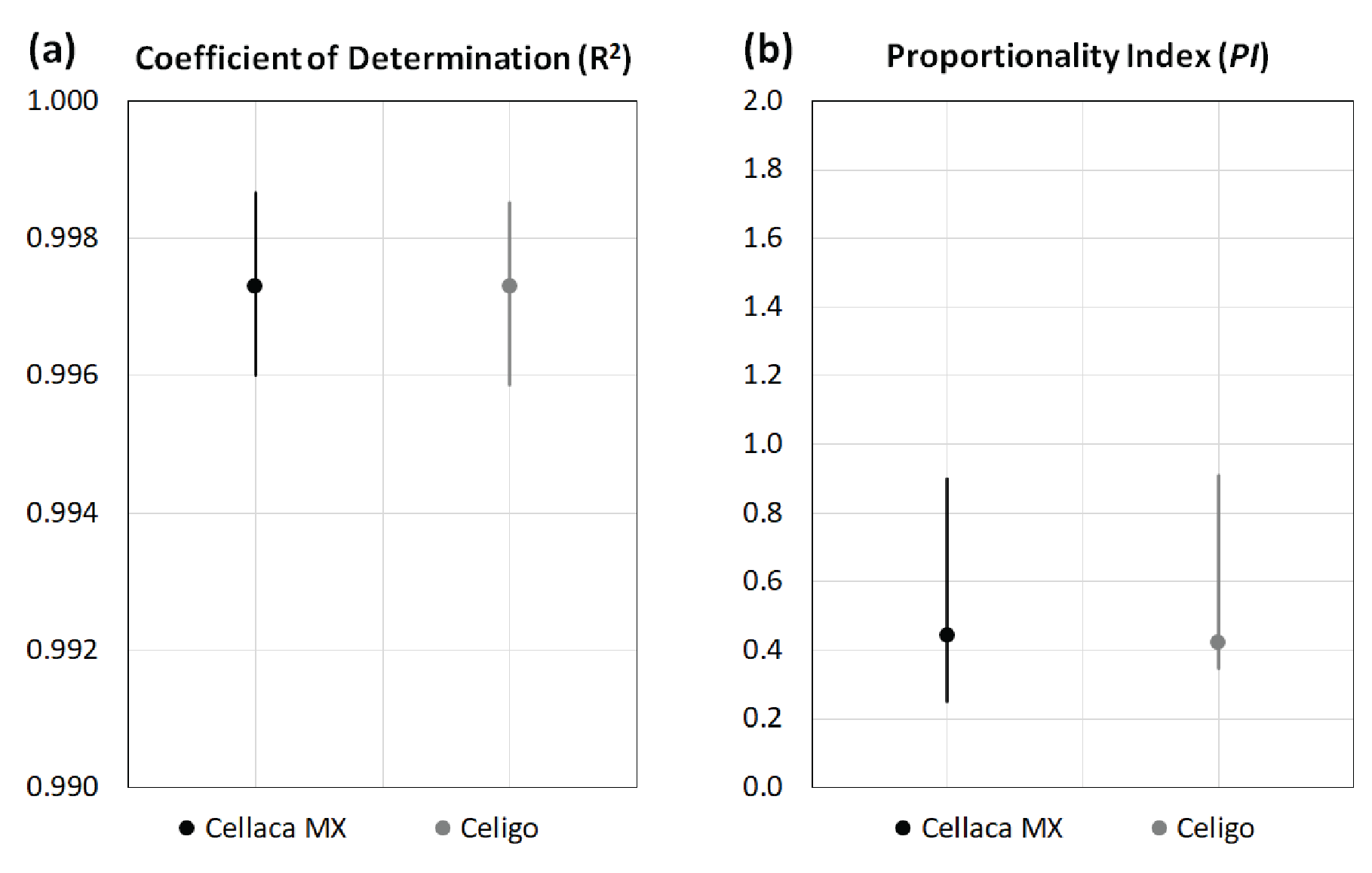
| Table 5 Calculated R-square and PI values for performance evaluation. | ||
| R-Square | PI | |
| Cellaca MX | 0.997 | 0.44 |
| Celigo | 0.997 | 0.42 |
| Significance | No | No |
Next, the Bland-Altman method is applied to compare the performance between Cellaca MX and Celigo cell counting methods. Figure 5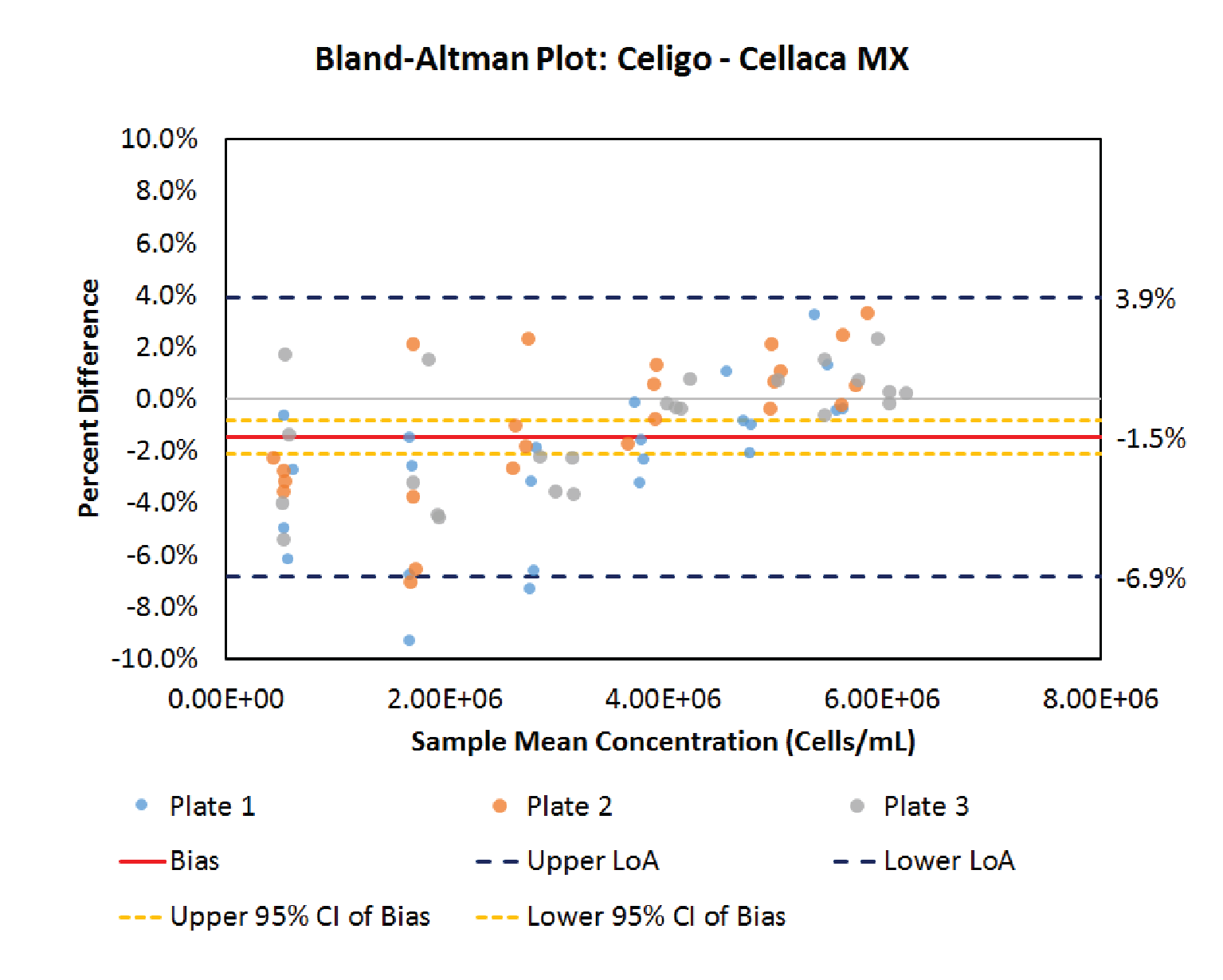
.5% lower than Cellaca MX in this cell counting method comparison. Since the value of 0 lies outside the confidence interval of the bias, it is concluded that the concentration difference observed between these two cell counting methods is significant (p < 0.05), despite being relatively small. The bias exhibits a slight dependence on cell concentration in this case. An additional paired measurement made using two methods under the same conditions would be expected to fall within the range defined by the LoA’s in approximately 95% of cases.
| Table 6 Bland-Altman comparative analysis results between Cellaca MX and Celigo. | ||
| Bias | Limit of agreement | CI of bias |
| -1.5% | -6.9% to 3.9% | -2.1% to -0.8% |
A summary of the cell counting performance evaluation and comparison results between Cellaca MX and Celigo using different combinations of targeted cell lines and staining solutions is shown in Table 7. Concentration biases are -2–6% for the paired cell counting methods in these four cell counting methods. Because each cell-stain combination was treated independently, the results were not combined, and the False Discovery Rate (FDR) or Family-Wise Error Rate (FWER) were not calculated. Overall, we have conducted multiple practical experiments following ISO Cell Counting Standard Part 2 with Celigo and Cellaca MX, and the results presented here are in the expected range of the cell counting measurement quality.
| Table 7 Summary table of cell counting performance evaluation and comparison results. | |||||||||||
| Cell line | Staining solution | Cellaca MX | Celigo | Bias | LoA | 95% CI of bias | Significance of bias | ||||
| R2 | Pooled CV range (%) | PI | R2 | Pooled CV range (%) | PI | ||||||
| Jurkat | Nuclear green | 0.998 | 3.8% to 6.1% | 0.30 | 0.997 | 3.7% to 7.4% | 0.37 | 3.4% | -6.0% to 12.8% | 2.3% to 4.5% | Y |
| Jurkat | AO | 0.997 | 2.2% to 7.5% | 0.44 | 0.997 | 1.8% to 7.6% | 0.42 | -1.5% | -6.9% to 3.9% | -2.1% to -0.8% | Y |
| CHO | Nuclear green | 0.998 | 3.4% to 5.8% | 0.40 | 0.999 | 2.7% to 6.8% | 0.35 | 5.6% | -3.4% to 14.6% | 4.5% to 6.7% | Y |
| CHO | AO | 0.998 | 2.7% to 7.0% | 0.44 | 0.996 | 2.7% to 6.4% | 0.35 | 5.1% | -2.4% to 12.5% | 4.2% to 5.9% | Y |
The ISO Cell Counting Standard Part 2 enables cell and gene therapy researchers to conduct experiments to evaluate and compare the quality of cell counting measurement processes. In this practical application of the standard, we demonstrated the evaluation of the Cellaca MX and Celigo using Jurkat and CHO cells tailored to the bioprocessing and cell therapy communities. The users of the ISO Cell Counting Standard Part 2 may utilize the protocol to evaluate one or more cell counting methods. For the evaluation of one method, one can perform the experiments to establish a baseline for each of the quality parameters, where this baseline can be further monitored with different operators, instruments, processes, etc. For comparison of cell counting methods, the quality parameters can be compared via bootstrap analysis or replicate studies, and the difference or bias between the methods can be determined using the Bland-Altman comparative analysis. It is also important to note that the quality parameters obtained from the ISO Cell Counting Standard Part 2 are specific to the measurement process evaluated (e.g. operators, instruments, cell sample properties, etc.), thus the robustness of the measurement process should also be evaluated in order to extend the findings of the study to similar measurement processes.
Frequently asked questions
- Can we reduce the number of replicate samples and measurements?
- Yes, to an extent; the minimum recommended experimental design consists of at least 4 target dilution fractions, 3 replicate samples, and 3 replicate measurements.
- Quality indicators from experimental designs that do not meet these recommendations should be interpreted with caution and may require additional studies to properly evaluate proportionality and precision.
- For example, if an experimental design has only 2 replicate measurements, the evaluation of CV directly from this experimental design and statistical analysis may not be appropriate, however, evaluation of proportionality can still be conducted.
- In this case, a second experiment with more replicate measurements of fewer samples and/or fewer dilution fractions may be conducted to more directly address precision of the method.
- What is the cell concentration range we should use?
- It should be fit-for-purpose for the typical range of cell concentrations you intend to evaluate for your cell type.
- Should we check the pipettors?
- Always use professionally calibrated pipettors
- Performing a check on pipettors will increase confidence in the results
- ISO 20391-2 also suggest an approach for generating measured dilution fractions, where the mass of solution pipetted while generating the fractions is used to calculate a more accurate measured dilution fraction value for use in the analysis of R2 and PI. In this case, small errors in pipetting can be accounted for in the proportional model fit.
- What can the results tell us?
- The quality indicators provide a means to quantify and compare the quality of cell counting methods based on principles that are fundamental to counting: precision and proportionality
- The ISO Counting Standard Part 2 analysis makes no assumptions about the true cell count and can make conclusions about method quality in the absence of a reference material or reference method.
- The Bland-Altman comparative analysis will indicate the percent difference between 2 methods.
- These approaches do not indicate or compare the accuracy of the cell counting methods.
- Cell counting method selection should be made based on the quality of the method and on what is fit-for-purpose for your measurement needs.
References
1. Boyiadzis MM, Dhodapkar MV, Brentjens RJ et al. Chimeric antigen receptor (CAR) T therapies for the treatment of hematologic malignancies: clinical perspective and significance. J. ImmunoTher. Cancer 2018; 6(137): 1–12. Crossref
2. Gonçalves GAR, Paiva RdMA. Gene therapy: advances, challenges and perspectives. Einstein 2017; 15(3): 369–75. Crossref
3. Huang R, Li X, He Y et al. Recent advances in CAR-T cell engineering. J. Hematol. Oncol. 2020; 13(86): 1–19. Crossref
4. Li Y, Huo Y, Yu L, Wang J. Quality Control and Nonclinical Research on CAR-T Cell Products: General Principles and Key Issues. Engineering 2019; 5: 122–31. Crossref
5. Seimetz D, Heller K, Richter J. Approval of First CAR-Ts: Have we Solved all Hurdles for ATMPs? Cell Med. 2019; 11: 1–16. Crossref
6. Nagata S, Hanayama R, Kawane K. Autoimmunity and the Clearance of Dead Cells. Cell 2010; 140(5): 619–30. Crossref
7. Rock KL, Kono H. The inflammatory response to cell death. Ann. Rev. Pathol. 2008; 3: 99–126. Crossref
8. Zhang Y, Chen X, Gueydan C, Han J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018; 28: 9–21. Crossref
9. 21st Century Cures. United States of America; 2016.
10. Arcidiacono JA, Bauer SR, Kaplan DS, Allocca CM, Sarkar S, Lin-Gibson S. FDA and NIST collaboration on standards development activities supporting innovation and translation of regenerative medicine products. Cytotherapy 2018; 20: 779–84. Crossref
11. Lin-Gibson S, Sarkar S, Elliott JT. Summary of the National Institute of Standards and Technology and US Food And Drug Administration cell counting workshop: Sharing practices in cell counting measurements. Cytotherapy 2018; 20(6): 785–95. Crossref
12. Biotechnology – Cell counting – Part 1: General guidance on cell counting methods. In: Standardization IOf, editor; 2018. Crossref
13. Biotechnology – Cell counting – Part 2: Experimental design and statistical analysis to quantify counting method performance. In: Standardization IOf, editor; 2019. Crossref
14. Validation of Analytical Procedures: Text and Methodology Q2(R1). In: Guideline IHT, editor; 2005. Crossref
15. Guidance for Industry: Process Validation: General Principles and Practices. In: Services USDoHaH, Administration FaD, (CDER) CfDEaR, (CBER) CfBEaR, (CVM) CfVM, editors; 2011. Crossref
16. Guidance for Industry: Analytical Procedures and Methods Validation for Drugs and Biologics. In: Services USDoHaH, Administration FaD, (CDER) CfDEaR, (CBER) CfBEaR, editors; 2015. Crossref
17. Sarkar S, Lund SP, Vyzasatya R, Vanguri P, Elliott JT, Plant AL, Lin-Gibson S. Evaluating the quality of a cell counting measurement process via a dilution series experimental design. Cytotherapy 2017; 19(12): 1509–21. Crossref
18. Sarkar S, Pierce L, Lin-Gibson S, Lund SP. Standards Landscape in Cell Counting: Implications for Cell & Gene Therapy. Cell Gene Ther. Ins. 2019; 5(1): 117–31. Crossref
19. Cohen J. The earth is round (p<.05). Am. Psychol. 1994; 49(12): 997–1003.
20. Altman DG, Bland JM. Measurement in Medicine: The Analysis of Method Comparison Studies. J. Royal Stat. Soc. Series D (The Statistician) 1983; 32(3): 307–17. Crossref
21. Bland JM, Altman DG. Statistical Methods For Assessing Agreement Between Two Methods of Clinical Measurement. Lancet 1986; 327(8476): 307–10. Crossref
22. Tholudur A, Giron L, Alam K et al. Comparing Automated and Manual Cell Counts for Cell Culture Applications. BioProcess Int. 2006. Crossref
23. Carkeet A. A Review of the Use of Confidence Intervals for Bland-Altman Limits of Agreement in Optometry and Vision Science. Optometry Vision Sci. 2020; 97(1): 3–8. Crossref
24. Carkeet A. Exact Parametric Confidence Intervals for Bland-Altman Limits of Agreement. Optometry Vision Sci. 2015; 92(3): e71–e80. Crossref
25. Abu-Arafeh A, Jordan H, Drummond G. Reporting of method comparison studies: a review of advice, an assessment of current practice, and specific suggestions for future reports. Br. J. Anaesthesia 2016; 117(5): 569–75. Crossref
26. Navidi W. Statistics for Engineers and Scientists. New York: McGraw-Hill; 2013. Crossref
27. Sokal RR, Rohlf FJ. Biometry. New York: W. H. Freeman and Company; 1969. Crossref
Affiliations
Yongyang Huang
Department of Advanced Technology R&D, Nexcelom Bioscience LLC, Lawrence, MA, USA
Jordan Bell
Department of Advanced Technology R&D, Nexcelom Bioscience LLC, Lawrence, MA, USA
Dmitry Kuksin
Department of Advanced Technology R&D, Nexcelom Bioscience LLC, Lawrence, MA, USA
Sumona Sarkar
Biosystems and Biomaterials Division, Materials Measurement Laboratory, National Institute of Standards and Technology, Gaithersburg, MD, USA
Laura T Pierce
Biosystems and Biomaterials Division, Materials Measurement Laboratory, National Institute of Standards and Technology, Gaithersburg, MD, USA
David Newton
Statistical Engineering Division, Information Technology Laboratory, National Institute of Standards and Technology, Boulder, CO, USA
Jean Qiu
Department of Advanced Technology R&D, Nexcelom Bioscience LLC, Lawrence, MA, USA
Leo Li-Ying Chan
Author for correspondence:
Department of Advanced Technology R&D, Nexcelom Bioscience LLC, Lawrence, MA, USA
Authorship & Conflict of Interest
Contributions: Leo Li-Ying Chan and Jean Qiu conceived the study and experimental design. Dmitry Kuksin performed the cell culture and provided the Jurkat and CHO-S cells for the experimentation. Jordan Bell and Yongyang Huang developed software application for the ISO Cell Counting Standards Part II parameters and the Bland-Altman analysis method. Yongyang Huang conducted the cell counting method performance evaluation and comparison experiments. Sumona Sarkar and Laura T Pierce contributed to the development of cell counting standardization, which this protocol was derived from. David Newton contributed to the development and description of the statistical analysis. Leo Li-Ying Chan, Yongyang Huang, Jordan Bell, Sumona Sarkar, and Laura T Pierce wrote the manuscript.
Acknowledgements: The authors would like to thank Pauline N. Mitchell for providing the art work of the Cell sample preparation and measurement process diagram.
Disclosure and potential conflicts of interest: Yongyang Huang, Jordan Bell, Dmitry Kuksin, Jean Qiu, and Leo Li-Ying Chan declare competing interests. The application of the cell counting method performance evaluation and comparison employed cell counting systems and reagents from Nexcelom Bioscience LLC.
NIST Copyright disclaimer: Official contribution of the National Institute of Standards and Technology; not subject to copyright in the United States.
NIST Commercial product disclaimer: Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Funding declaration: The authors received no financial support for the research, authorship and/or publication of this article.
Article & copyright information
Copyright: Published by Cell and Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0 which allows anyone to copy, distribute, and transmit the article provided it is properly attributed in the manner specified below. No commercial use without permission.
Attribution: Copyright © 2021 Nexcelom Bioscience. Published by Cell and Gene Therapy Insights under Creative Commons License Deed CC BY NC ND 4.0.
Article source: Invited; externally peer reviewed.
Submitted for peer review: Jul 2 2021; Revised manuscript received: Aug 19 2021; Publication date: Sep 15 2021.

